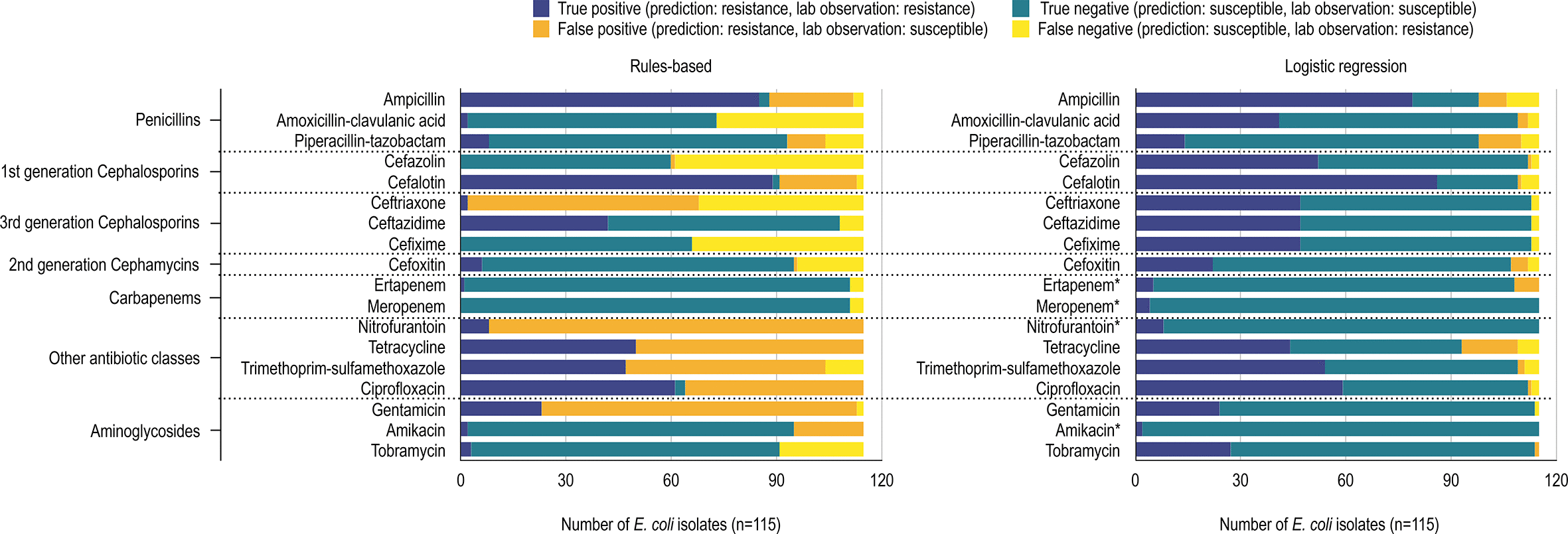
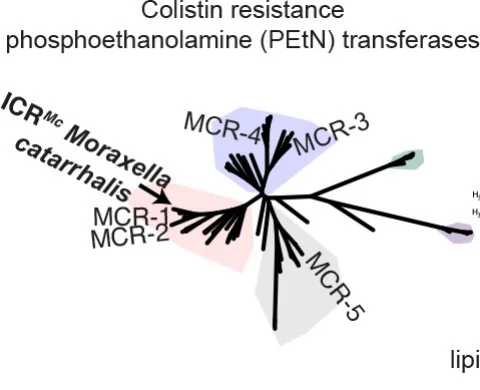
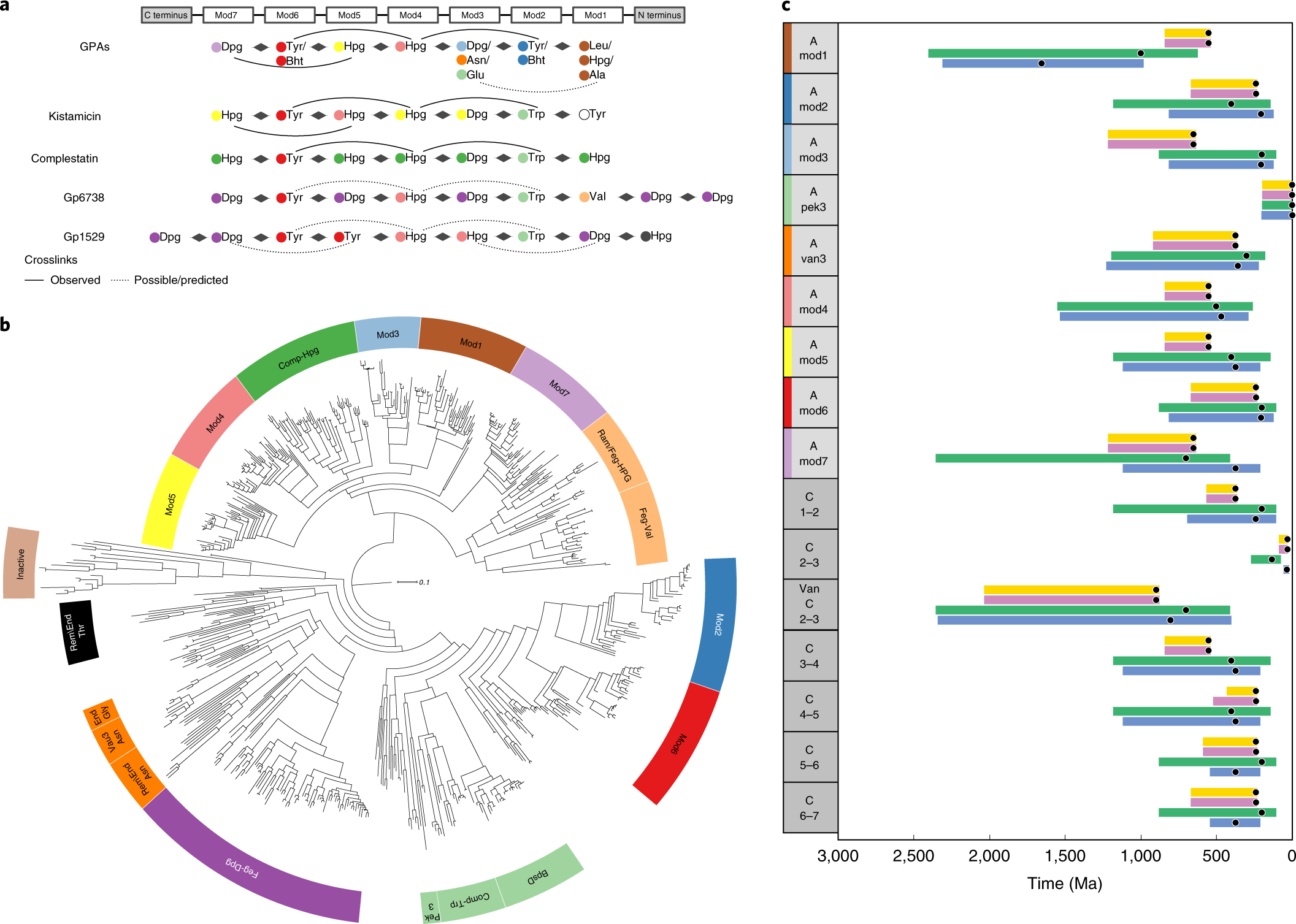
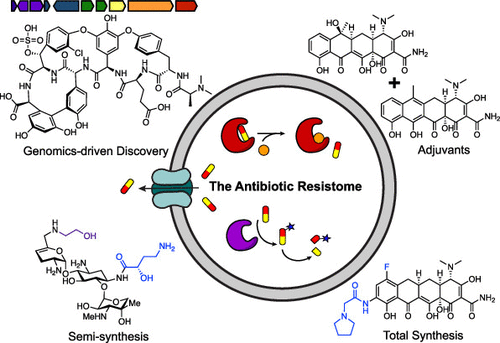
Antibiotic Resistance
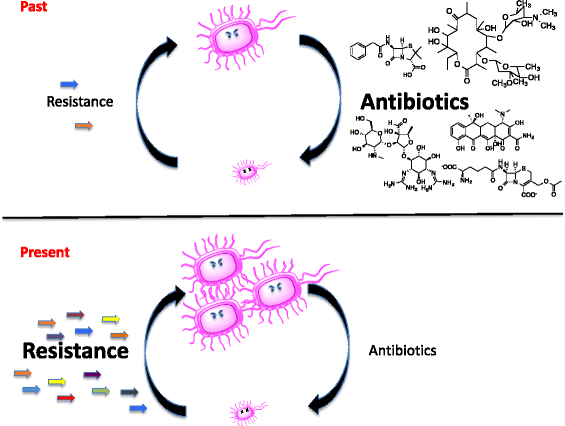
Resistance to antibiotics is a clinical crisis of global proportion. The ability of microbes to evade antibiotics and the lack of new compounds emerging from the pharmaceutical sector means that we must have a thorough understanding of the mechanisms of resistance and its evolution. We are using a strategy termed Genomic Enzymology to study antibiotic resistance in our lab. In this approach, predicted antibiotic resistance genes are identified from available genome sequences or by direct cloning of new resistance elements based on phenotype. This is followed by rigorous structure/function analysis to understand the molecular details of resistance.
Mechanisms of antibiotic resistance
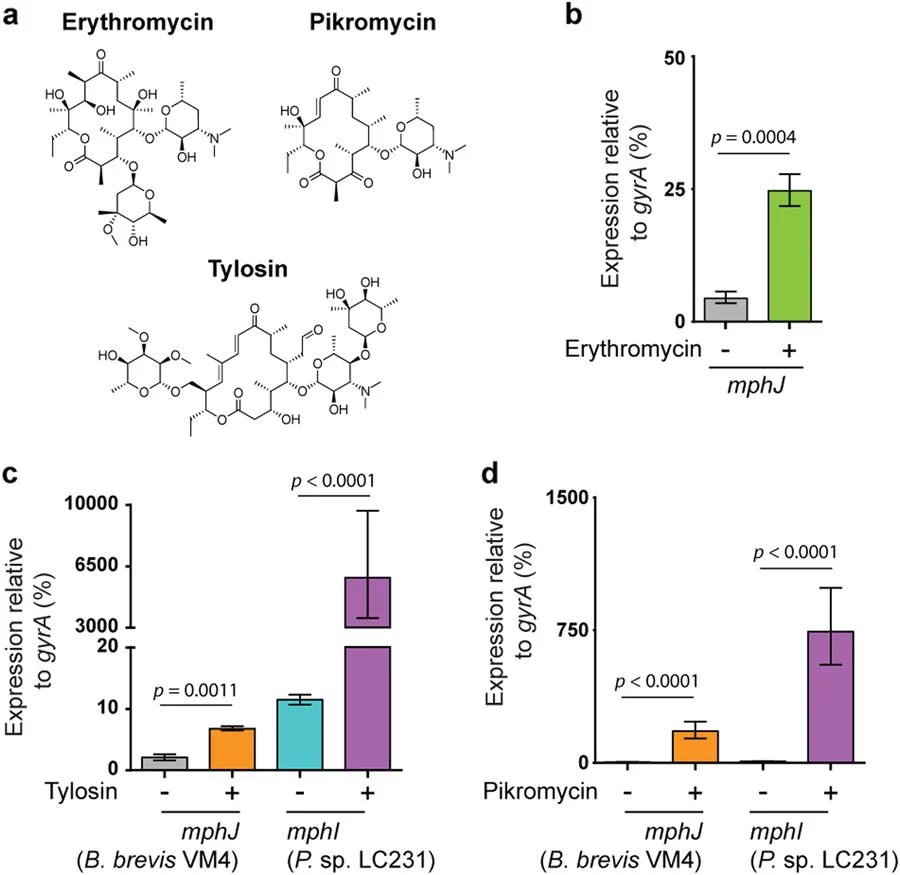
We are studying the mechanisms of resistance to several classes of antibiotics including the aminoglycosides, glycopeptides, tetracyclines, streptogramins, lipopeptides, and rifamycins. This includes understanding the detailed molecular mechanisms and the structures of antibiotic resistance proteins. In particular, we are focused on enzymes that inactivate the compounds or bypass the molecular target. Our goals are to fully understand the molecular details of resistance using protein structure and function approaches. With this information, we can begin to strategize around methods to block or otherwise avoid these resistance mechanisms.
Selected Publications
Authors: Surette MD, Waglechner N, Koteva K, Wright GD.
Reference: Mol Cell.
DOI: 10.1016/j.molcel.2022.06.019.
PMID: 35907401
Authors: Stogios PJ, Bordeleau E, Xu Z, Skarina T, Evdokimova E, Chou S, Diorio-Toth L, D'Souza AW, Patel S, Dantas G, Wright GD, Savchenko A.
Reference: Commun Biol.
DOI: 10.1038/s42003-022-03219-w.
PMID: 35338238.
Authors: Zubyk HL, Wright GD.
Reference: Antimicrob Agents Chemother.
DOI: 10.1128/AAC.00773-21.
PMID: 34001507.
Authors: Bordeleau E, Stogios PJ, Evdokimova E, Koteva K, Savchenko A, Wright GD.
Reference: mBio
DOI: 10.1128/mBio.02705-20
PMID: 33563840
Authors: Tsang KK, Maguire F, Zubyk HL, Chou S, Edalatmand A, Wright GD, Beiko RG, McArthur AG.
Reference: Microb Genom
DOI: 10.1099/mgen.0.000500
PMID: 33416461
Authors: Schaenzer AJ and Wright GD
Reference: Trends Mol Med.
DOI: 10.1016/j.molmed.2020.05.001
PMID: 32493628
Authors: Zubyk HL, Cox G and Wright GD
Reference: J Vis Exp.
DOI: 10.3791/60536
PMID: 31680676
Authors: Thomy D, Culp E, Adamek M, Cheng EY, Ziemert N, Wright GD, Sass P and Brötz-Oesterhelt H.
Reference: Appl Environ Microbiol.
DOI: 10.1128/AEM.01292-19
PMID: 31399403
Authors: Stogios PJ, Cox G, Zubyk HL, Evdokimova E, Wawrzak Z, Wright GD, Savchenko A
Reference: ACS Chemical Biology 2018 13 (5), 1322-1332
DOI: 10.1021/acschembio.8b00116
PMID: 29631403
Authors: Koteva K, Cox G, Kelso JK, Surette MD, Zubyk HL, Ejim L, Stogios P, Savchenko A, Sørensen, Wright GD
Reference: Cell Chemical Biology 2018 Jan 26. pii: S2451-9456(18)30031-X
DOI: 10.1016/j.chembiol.2018.01.009.
PMID: 29398560
Origins and evolution of antibiotic resistance
Where does antibiotic resistance come from?
Understanding this process can help track the emergence and spread of resistance mechanisms and help us to comprehend the forces at work in selection for resistance. We approach this in two ways. First, we are searching the genomes of bacteria for genes that encode homologues of resistance enzymes and studying the enzymology of these to assess their fitness as antibiotic inactivators. Second, we are actively screening environmental organisms for novel resistance phenotypes and identifying new mechanisms. This includes collecting organisms from diverse environments and studying the molecular mechanism of resistance.
Selected Publications
Authors: Waglechner N, Culp EJ, Wright GD.
Reference: EcoSal Plus.
DOI: doi: 10.1128/ecosalplus.ESP-0027-2020.
PMID: 33734062.
Authors: Waglechner N, McArthur AG, and Wright GD
Reference: Nat Microbiol.
DOI: 10.1038/s41564-019-0531-5
PMID: 31406334
Authors: Pawlowski AC, Stogios PJ, Koteva K, Skarina T, Evdokimova E, Savchenko A, Wright GD
Reference: Nature Communications 2018 ;9(1):112
DOI: 10.1038/s41467-017-02680-0.
PMID: 29317655
Authors: Pawlowski AC, Wang W, Koteva K, Barton HA, McArthur AG, Wright GD.
Reference: Nat Commun. 2016 Dec 8;7:13803. doi: 10.1038/ncomms13803.
PMID: 27929110
Authors: Vanessa M D’Costa, Christine E King, Lindsay Kalan, Mariya Morar, Wilson WL Sung, Carsten Schwarz, Duane Froese, Grant Zazula, Fabrice Calmels, Regis Debruyne, G Brian Golding, Hendrik N Poinar, Gerard D Wright
Reference: Nature. 2011 Aug 31;477(7365):457-61. doi: 10.1038/nature10388.
PMID: 21881561
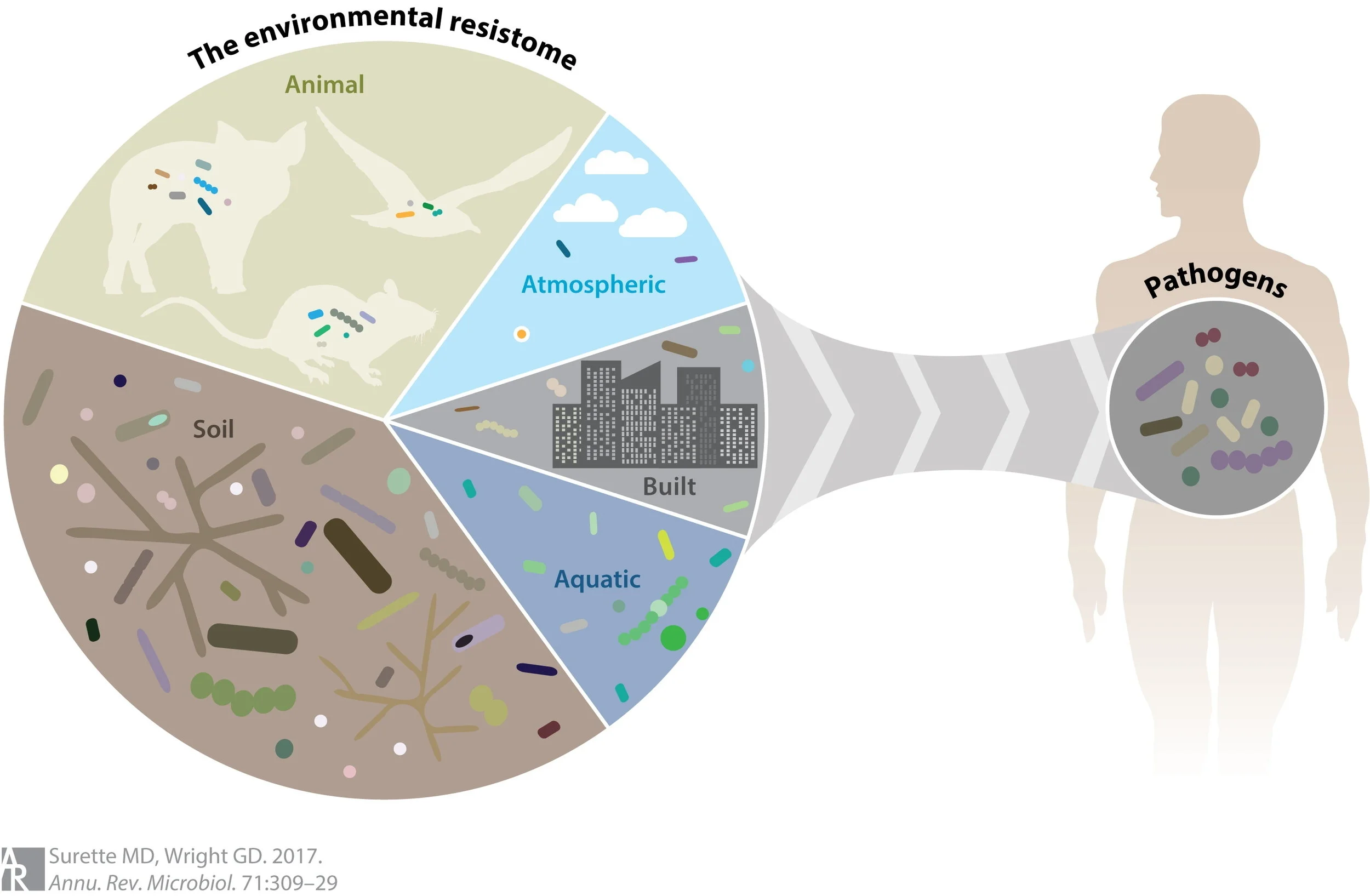
The Antibiotic Resistome: A framework to understand the evolution of resistance
We have termed the collection of all antibiotic resistance genes the “Antibiotic Resistome”. The resistome includes not only the resistance elements associated with pathogens, but also those genes in environmental organisms not usually associated with disease. Furthermore, genes that encode proteins that serve as ancestors of resistance, which we call proto-resistance genes in analogy to the cancer field.
Selected Publications
Authors: Guitor AK, Yousuf EI, Raphenya AR, Hutton EK, Morrison KM, McArthur AG, Wright GD, Stearns JC.
Reference: Microbiome.
DOI: 10.1186/s40168-022-01327-7.
PMID: PMC9414150
Authors: Surette MD, Spanogiannopoulos P, Wright GD.
Reference: Accounts of Chemical Research
DOI: 10.1021/acs.accounts.1c00048
PMID: 33877820.
Authors: Hobson C, Chan AN, Wright GD.
Reference: Chem Rev.
DOI: doi: 10.1021/acs.chemrev.0c01214.
PMID: 33606500.
Authors: Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG.
Reference: Nucleic Acids Res.
DOI: 10.1093/nar/gkz935
PMID: 31665441
Authors: Guitor AK, Raphenya AR, Klunk J, Kuch M, Alcock B, Surette MG, McArthur AG, Poinar HN and Wright GD
Reference: Antimicrob Agents Chemother.
DOI: 10.1128/AAC.01324-19
PMID: 31611361
Authors: Wright GD
Reference: Curr Opin Microbiol.
DOI: 10.1016/j.mib.2019.06.005
PMID: 31330416
Authors: Pawlowski AC, Westman EL, Koteva K, Waglechner N, Wright GD
Reference: ISME vol 12, 885–897
DOI: 10.1038/s41396-017-0017-5
PMID: 29259290
Authors: MD Surette and GD Wright
Reference: Annual Review of Microbiology Vol. 71:309-329
PMID: 28657887
Authors: N Waglechner and GD Wright
Reference: BMC Biology. 2017 Sep 15;15:84. doi.org/10.1186/s12915-017-0423-1
PMID: 28915805
Authors: Julie A Perry, Gerard D Wright
Reference: Bioessays. 2014 Dec;36(12):1179-84. doi: 10.1002/bies.201400128.
PMID: 25213620